An international team of researchers has unveiled a groundbreaking technique that promises to expedite drug design aimed at treating diseases linked to ion channels. This innovative method, developed by the Institute of Chemical Research, a collaboration between the University of Seville and the Spanish National Research Council, was published in the Journal of the American Chemical Society on November 15, 2025.
Ion channels are specific proteins located in cell membranes that play a crucial role in regulating the flow of ions into and out of cells. These proteins are implicated in various medical conditions, including psychiatric disorders and several cancers, making them important targets for therapeutic intervention.
Revolutionizing Drug Interaction Studies
Traditionally, studying drug interactions with ion channels required isolating these proteins, a method that is not only complex but also risks altering their behavior. According to Jesús Angulo, a researcher at the Institute of Chemical Research, the new technique leverages nuclear magnetic resonance to observe these interactions within living cells. This approach yields more biologically relevant data, enhancing the understanding of how drugs affect these critical proteins.
The innovative technique is not just faster but also more cost-effective and simpler. It eliminates the need for complicated preliminary steps such as protein purification. Experiments can now be conducted in less than an hour, significantly reducing the time required for drug design studies.
“This method could redefine the standard for structure-activity studies, which focus on the relationship between a molecule’s chemical structure and its pharmacological effects,” stated Leanne Stokes from the University of East Anglia.
Applications in Diverse Medical Fields
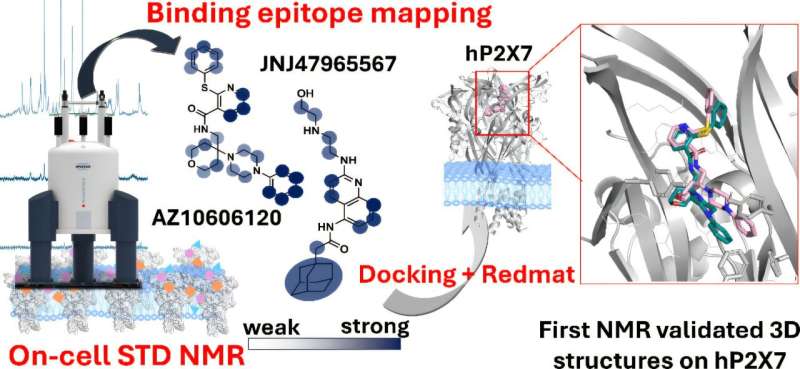
The researchers have already tested their technique on P2X7 receptors, which are potential therapeutic targets for conditions including depression, certain autism spectrum disorders, and various cancers. Serena Monaco, a researcher at the Quadram Institute, explained that the technique enables scientists to pinpoint precisely which parts of a drug interact with the target protein. This information is vital for optimizing drug interactions and developing more effective treatments.
Furthermore, the research team utilized software developed at IIQ-CSIC-US to integrate their experimental findings with three-dimensional models of drug-receptor binding created through bioinformatics. This integration allows for validation of which computer-generated models align with laboratory observations.
Angulo likened the interaction between a drug and its target protein to a lock and key mechanism. “The membrane protein acts as the lock, and our drug is the key. It is essential not only to find the right key but also to determine the best way to insert it to achieve effective activation,” he explained.
The advent of this technique marks a significant advancement in pharmacological studies, particularly in the design of new drugs targeting membrane proteins. “Validating three-dimensional computer models on living cells represents a new paradigm in drug development,” Angulo concluded.
The implications of this research extend across various medical fields, potentially paving the way for novel treatments in neurological, cardiovascular, metabolic, and oncological diseases. With this new tool, the pharmaceutical industry may see accelerated progress in addressing complex health challenges.
For further details, refer to the full study: Serena Monaco et al, “On-Cell Saturation Transfer Difference NMR Spectroscopy on Ion Channels: Characterizing Negative Allosteric Modulator Binding Interactions of P2X7,” published in the Journal of the American Chemical Society, DOI: 10.1021/jacs.5c02985.